A Personal Project with Sleep Data
I have always been fascinated by the process of planning, creating, and maintaining a backend database that can be used to hold data for analysis. That’s why I wanted to start my own project, using personal data that I have a good understanding of. I considered using information from my computer, phone, and apps, but they all seemed too vague. That’s when I realized that my CPAP machine, which I’ve been using since October 2022, was the perfect choice.
The CPAP machine collects various data points while I sleep, and I knew that this data would be valuable in providing insights into my daily life. The first step was to extract the data into a usable format.
Extracting and Cleaning the Data
Locating the Data
The machine I use, the Resvent iBreeze CPAP, comes with an app called iMatrix that allows me to view my historical sleep data. The data is uploaded to a Resvent server via WiFi, but is also stored on an SD card.
I wanted to find a way to use this data myself, so I started searching for a way to extract it. I found that tools like OSCAR couldn’t read the data from the Resvent iBreeze machine, but I was able to download the iMatrix software and upload the data from the SD card into it. This created a SQLite database that I could use to delve deeper into my sleep patterns.
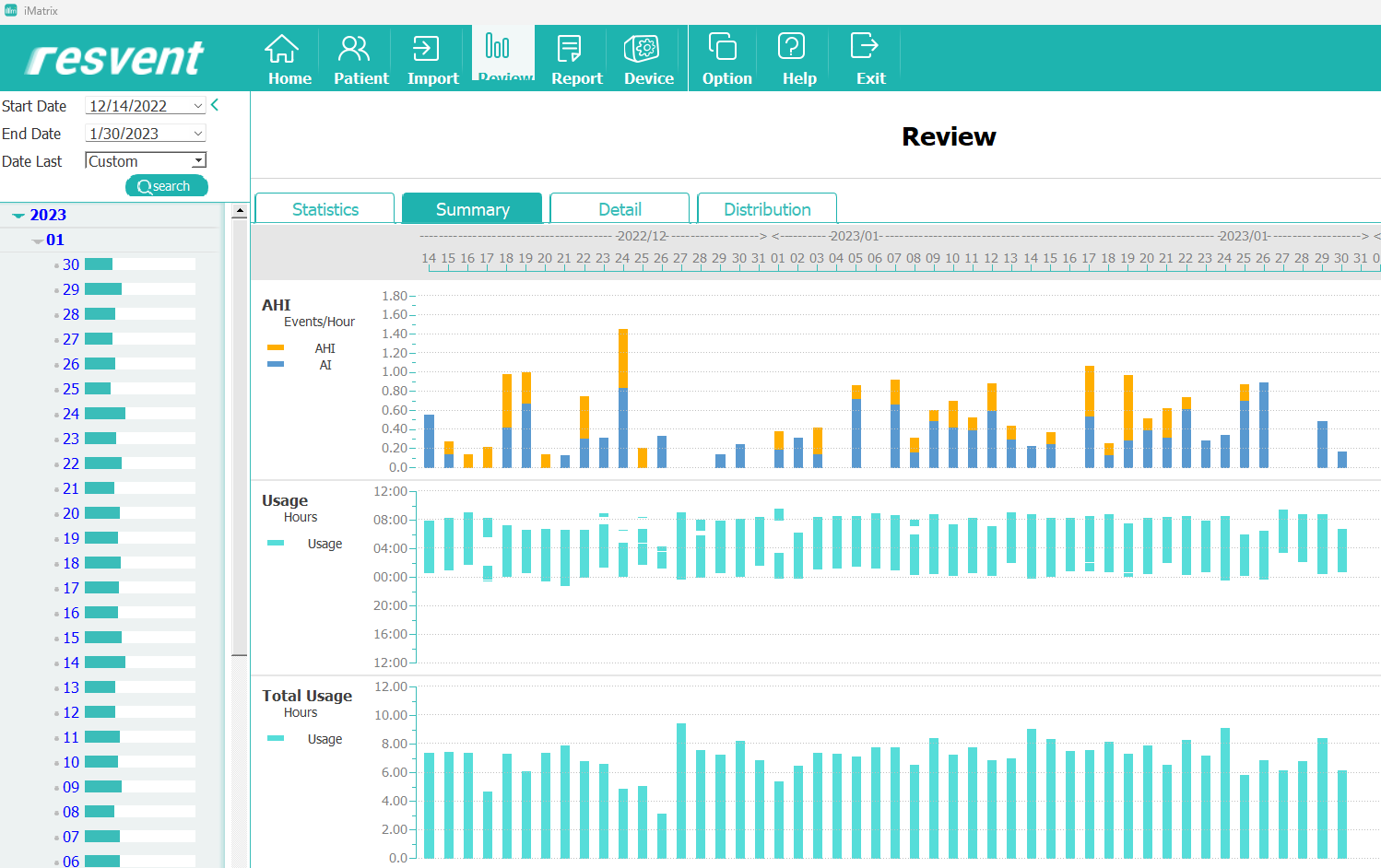
iMatrix graphs
Although the information from iMatrix was useful, I wanted to find a way to manipulate the data myself and bring in additional outside data. That’s when I discovered a Resvent_iBreeze_Data_Puller open source repo on GitHub. The python scripts from this repository, built by @Ryush806, allowed me to pull and process the data from the SD card.
Setting Up a Database
Before I could start pulling the data off the SD card, I needed to set up a database to store everything. I decided the database should be easy to configure, free, and self-hostable. I chose to use Postgres, as I was familiar with it and comfortable working with it and pgAdmin to set up the necessary tables. I created a CPAP database for this project and used DB Browser to read the SQLite database created by iMatrix.
After spending some time exploring the SQLite database and reviewing the code in the Resvent_iBreeze_Data_Puller repository, I was ready to start pulling the data.
Data Extraction
The scripts from the Resvent_iBreeze_Data_Puller repository were very thorough and allowed for multiple types of data extraction. The detailsPuller.py script allowed for the extraction of timestamped data collected by the CPAP machine every 2 seconds, including Pressure, IPAP, EPAP, Leak, Vt, MV, RR, TI, and IE data. The eventPuller.py provided timestamped information on significant events such as snores, flow limitations, or obstructive apneas.
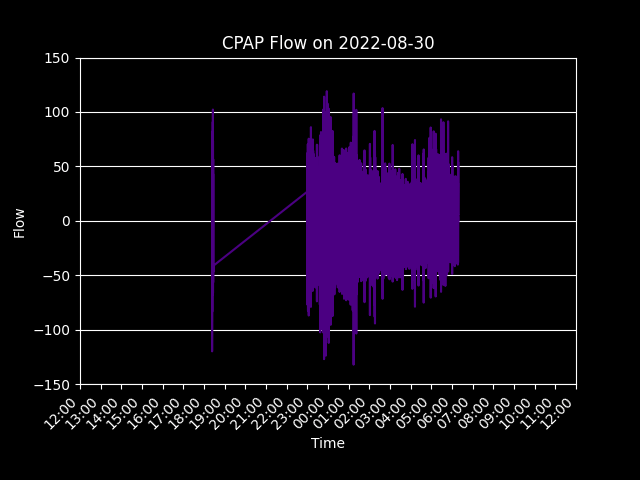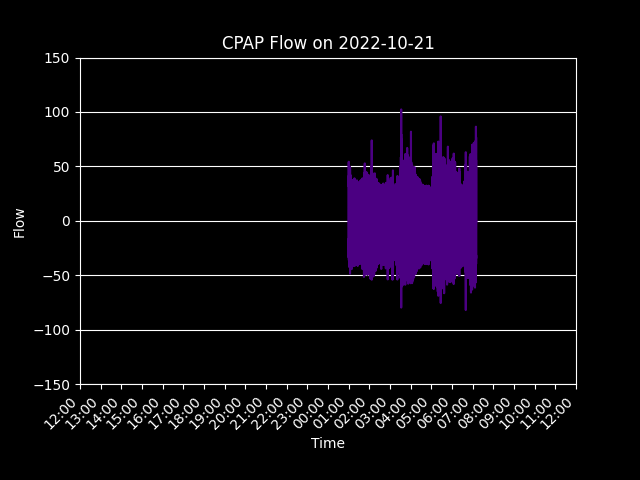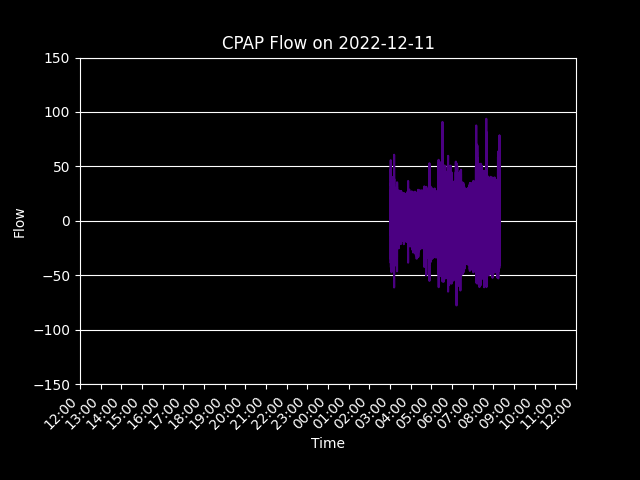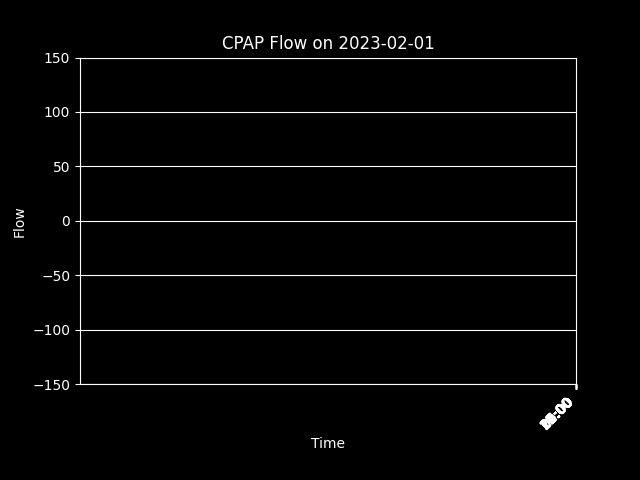
The wavePuller.py script allowed for the extraction of all pressure and flow data, which was collected by the machine every 20 milliseconds and stored as binary data. This generated two large CSV files, one for flow data and one for pressure data, each approximately 5 GB in size. While this data was not as crucial as the data extracted by the detailsPuller.py script, the source data was still desired and provided an opportunity to work with large amounts of data. When the flow and pressure data was uploaded to the Postgres database, it came out to be 98,246,000 rows long.
Data Loading and Cleansing
The next step was to load the extracted data into the Postgres database. This was done using the sqlalchemy library in Python. I utilized the create_engine and to_sql commands to load the data into the tables.
import pandas as pd
from sqlalchemy import create_engine
engine = create_engine('postgresql://<username>:<password>@localhost:5432/<dbname>')
df=pd.read_csv('E:\GitHub\cpap\wave\<csvname>.csv', low_memory=False)
df.to_sql('<tablename>', engine)
This code worked great to upload the csv data to my local database. However, I realized that sqlalchemy tables did not have the correct data types attributed to the columns it automatically created. I had to go through all the tables and correctly identify and assign the correct data types to them.
--- set timestamp
ALTER TABLE "be_waveFlowTable"
ALTER COLUMN timestamp SET DATA TYPE timestamp
USING to_timestamp(timestamp, 'YYYY-MM-DD HH24:MI:SS.MS');
--- set date
ALTER TABLE "be_waveFlowTable"
ALTER COLUMN date SET DATA TYPE date
USING to_date(date, 'yyyy-mm-dd');
--- set pressure and flow columns to numbers
ALTER TABLE "be_waveFlowTable"
ALTER COLUMN "Flow" SET DATA TYPE numeric USING "Flow"::numeric;
Once these steps were completed, the data extraction and loading process was successful. I was now able to work with my sleep data and start analyzing it. The data provided a lot of valuable insights, and I was excited to dive deeper.
Having the data on the server allowed for me to start making insightful graphs like the one below. This GIF shows my breathing flow as I use the machine each night.